Research
We are currently working on following research topics.
- Regulation of Chromatin Structure
- Nucleolar Structure and Functions
- Regulation of Gene Expression Program
- Proteomic Approach to Signal Transduction Pathways
- Drug Development for Anti-Infectious Diseases
Regulation of chromatin structure
Long human genomic DNA (about 2 m in human cells) is packed in the cell nucleus with 10 micrometers in diameter. To allow this compaction, chromatin structure plays a critical role. Nucleosome consisting of 4 core histones, H2A, H2B, H3, and H4 and 147 base pairs of double stranded DNA is a repeating unit of chromatin. Linker histones binds to the linker region of DNA between nucleosomes and help to form high order chromatin structure. We have identified and studied the functions of a group of proteins termed histone chaperones, which assemble and disassemble chromatin structure to allow gene expression and genomic DNA replication. Our research goal is to elucidate the molecular mechanisms by which chromatin structure is assembled and disassembled by focusing on the interaction between histone chaperones and histones including histone variants. In addition, histone chaperons identified in our lab are involved in sperm chromatin dynamics during spermatogenesis and fertilization. We are also interested in clarifying the molecular mechanism by which sperm chromatin is reconstructed during spermatogenesis and fertilization.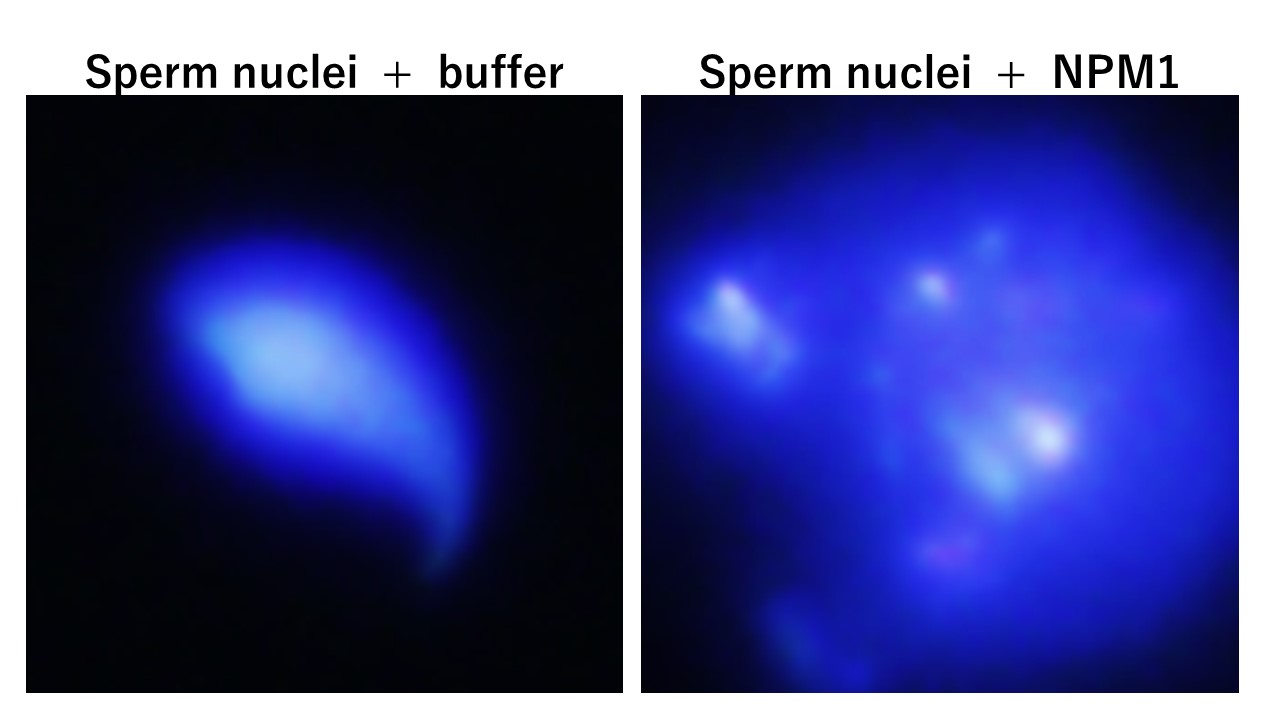
Nucleolar structure and functions
Nucleolus is well observed in growing cells under light microscope and its primary function is to synthesize ribosomes, which are protein translation machines. Ribosome is consisting of 4 ribosomal RNA (rRNAs) and about 80 ribosomal proteins. Among 4 rRNAs, 18S, 5.8S, and 28S rRNAs are synthesized in the nucleolus by RNA polymerase I as a single precursor RNA. Long precursor rRNA is successively processed by various proteins and RNAs. Ribosomal proteins are assembled on the rRNAs co-transcriptionally or after appropriate processing, although the detailed mechanism of the ribosomal protein assembly process is unknown. Given that growing cells consume a huge number of ribosomes, ribosome synthesis activity is correlated with the cell growth rate. Therefore, highly malignant cells show abnormally large nucleolus and the nucleolar abnormalities are sometimes used as a diagnostic marker of transformed cells. In addition, it should be noted that ribosome biogenesis is the most energy consuming reaction in the cells and therefore the ribosome biogenesis activity is very sensitive to the cellular energy status. The functions of the nucleolus to sense the energy level of the cells is also closely related to cellular aging. We are currently trying to elucidate the molecular mechanism of the nucleolar formation to contribute to understand cellular aging.
Regulation of gene expression program
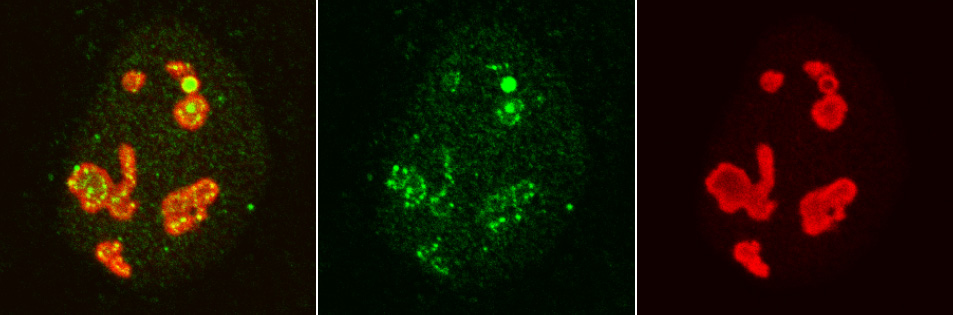
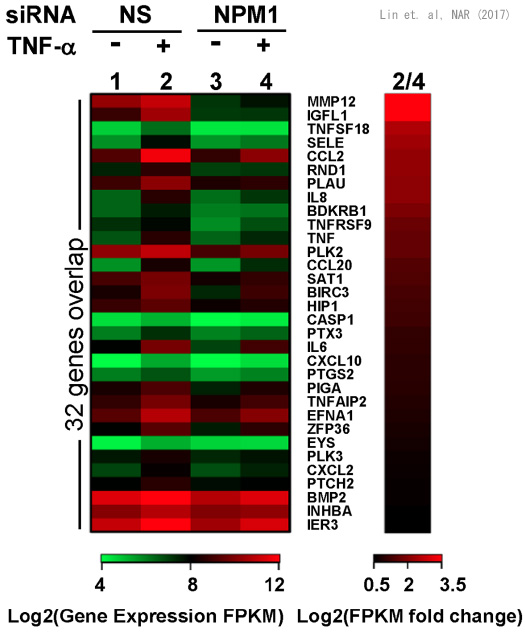
We have identified histone chaperones that regulate the chromatin structure. Recently, we showed that histone chaperone NPM1 directly binds to a transcription factor NFkB and regulates its DNA binding activity. Because NFkB is a central transcription factor in inflammatory responses and immune responses, it is presumed that NPM1 is also involved in these processes. In addition to NFkB, our research has suggested that other transcription factors are under control of NPM1. We are currently studying the interaction between histone chaperones and transcription factor, to elucidate the biological significance of this interaction.
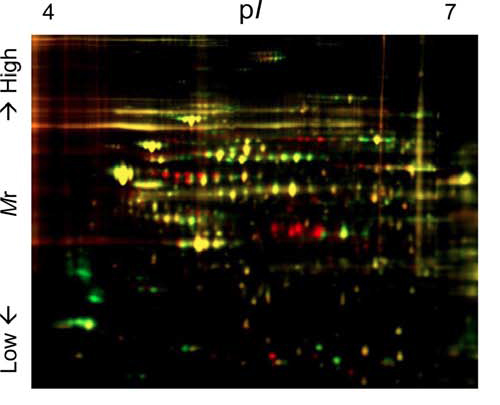
Proteomic approach to signal transduction pathways
It has been well established that abnormal signaling causes various diseases
including cancer and metabolic syndromes. Therefore, it is very important
to understand how the extracellular stimuli are properly transduced to
cells. To uncover the full view of the signal transduction pathway, we
are focusing on a well-established protein post-translational modification,
phosphorylation and de-phosphorylation. By combining 2-dimentional gel
electrophoresis and mass spectrometry analyses, we are trying to visualize
the signal transduction pathways.
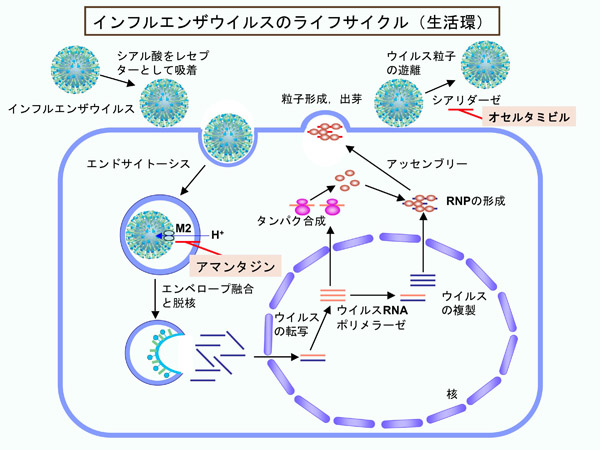
Development of drugs for anti-infectious diseases
We are trying to develop novel anti-influenza virus drugs in collaboration with the lab of Professor Tomoda in Kitasato University. Influenza viruses uses the Cap structure from host mRNA as a primer of viral mRNA synthesis. The viral RNA-dependent RNA polymerase complex mediates to endonucleoritically cleave mRNAs containing the Cap structure (Cap-snatching). Because host cells do not have this endonuclease activity, the viral nuclease activity is a good target for drug discovery. We have originally established the assay system to monitor the Cap-snatching by the viral RNA polymerase complex. Using this assay system, we are now trying to identify molecules that inhibit the viral mRNA synthesis.